About
I'm a scientist currently based in San Francisco.
I co-founded NewLimit to
develop new therapies based on reprogramming to reduce the burden of age-related disease.
Before that, I was in Seattle at AWS working new and diverse
technologies to
make the world a better place.
Before that, I was also in Seattle at the Allen Institute
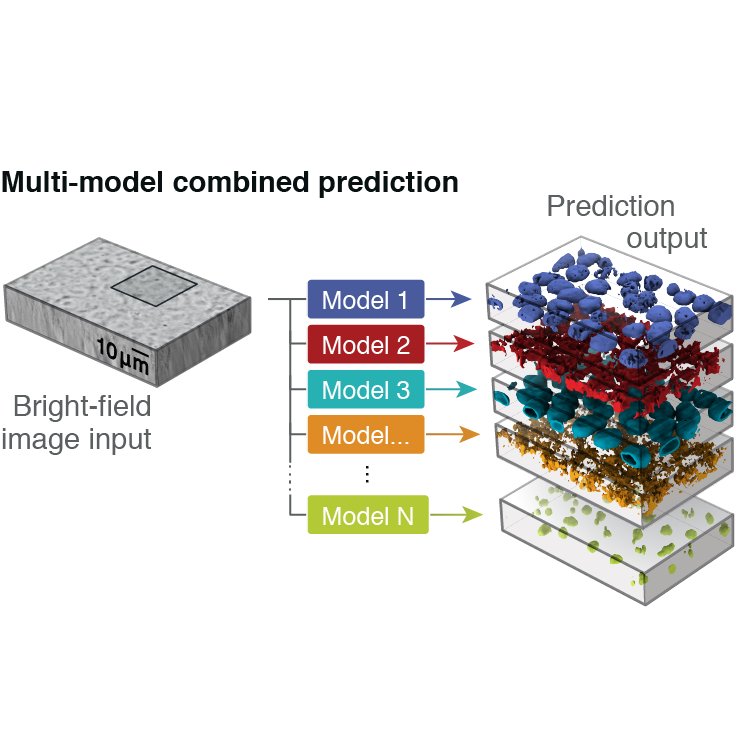
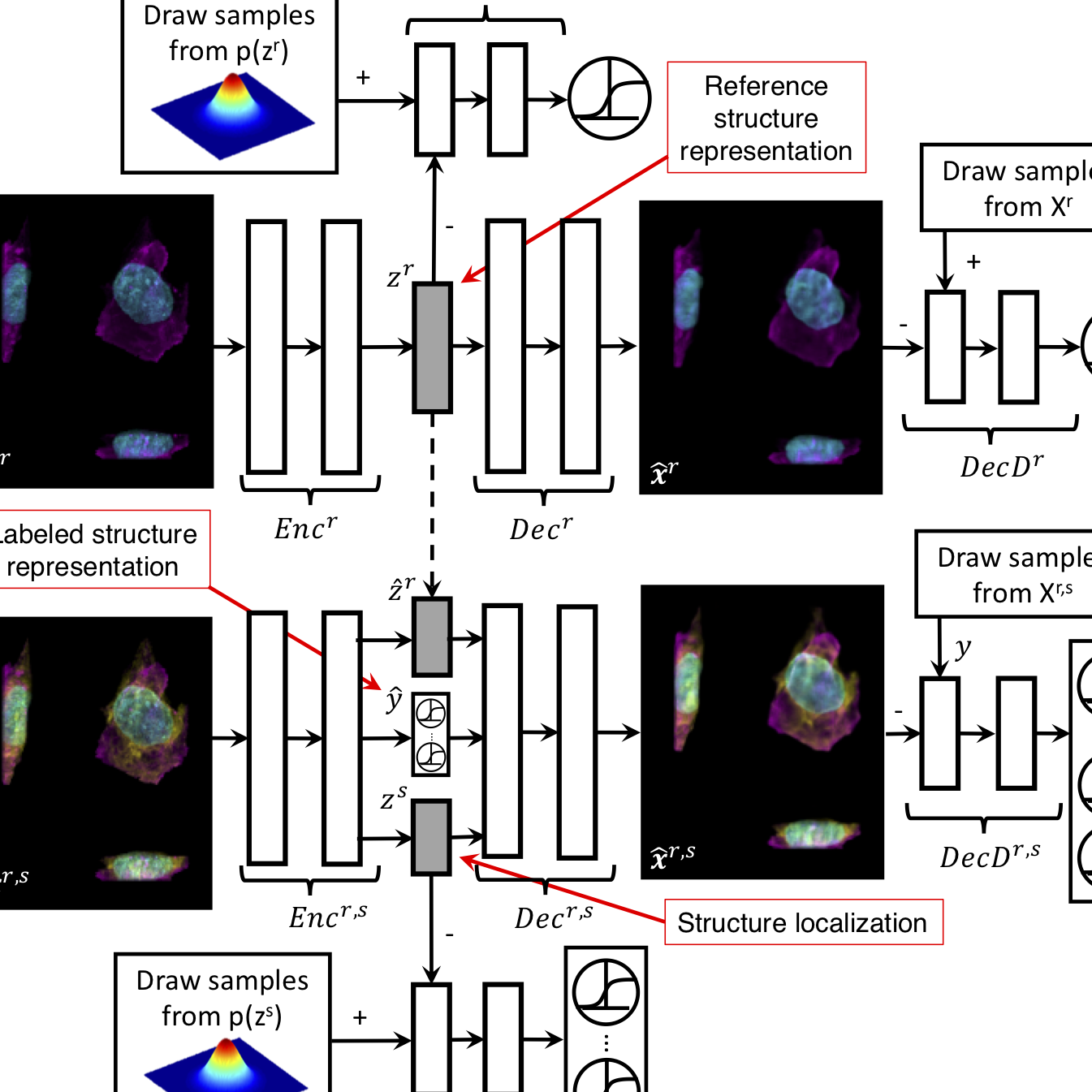
for Cell Science where I built predicitve models to estimate the outcome of new experiments and
describe how cells change their organization under different conditions.
Before that, I was in grad school at Carnegie Mellon University
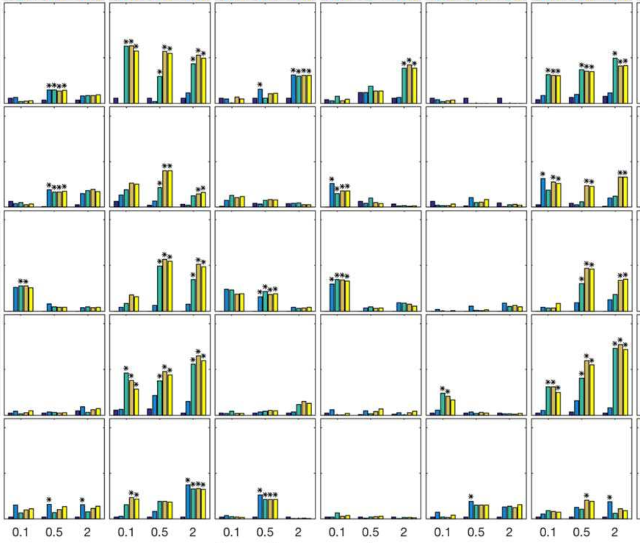
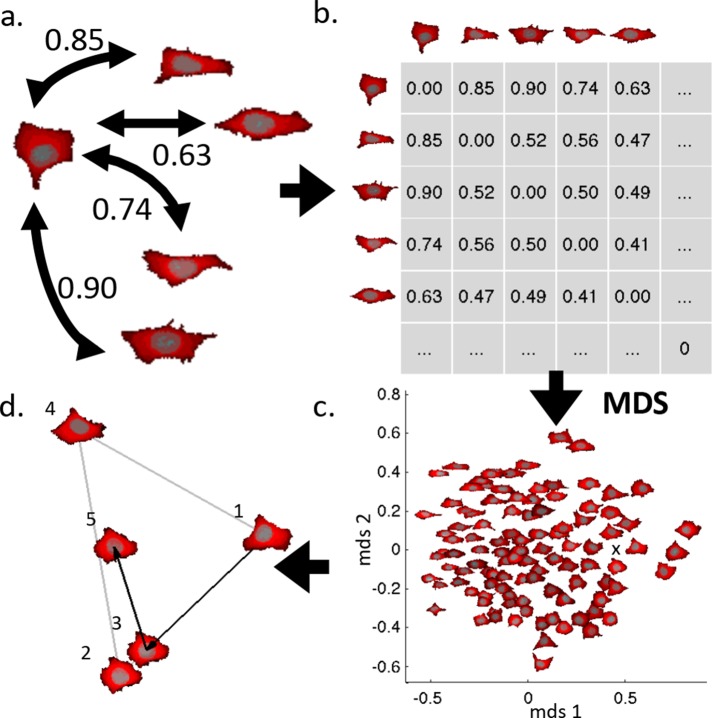
as a student of Robert Murphy. My work
focused on applying generative models to determine how cells respond to perturbations.
Even before that, I developed regulated document management software at Amgen.